The overarching goals of work in the Oldham Lab are to:
i) understand the molecular basis of cellular identity in the human brain in health and disease;
ii) develop novel treatments for brain tumors;
iii) change the way that scientists communicate around data analysis.
The human brain is an extraordinarily complex, heterogeneous structure, comprised of diverse cell types whose molecular and functional identities are poorly understood. Because gene expression lies at the root of cellular identity, much of our work focuses on understanding the organization of the human brain transcriptome. To this end, we generate and analyze vast quantities of gene expression data from human brain samples and use custom computational pipelines to identify and compare patterns of gene activity across many independent datasets.
By analyzing transcriptional signatures of malignant clones and nonmalignant cell types from human tumor samples, we are expanding the therapeutic search space for human cancers beyond the mutated gene products that often initiate or maintain tumor growth. We are particularly interested in identifying and exploiting molecular phenotypes that distinguish tumor vasculature from surrounding normal vasculature. Through integrative analysis of thousands of normal human brain and malignant glioma samples, we have identified highly reproducible markers of glioma vasculature and are performing functional studies to evaluate and prioritize these candidates as novel therapeutic targets.
Through our meta-analyses of published datasets, we have also identified many factors that contribute to irreproducible research, which imposes staggering societal costs and impedes efforts to develop novel and effective therapies for human diseases. To address these challenges, we are developing a cloud-based platform for storing, retrieving, and representing data, metadata, and code related to our lab’s computational research activities. By structuring biomedical data and metadata in a consistent fashion, we seek to create a precise foundation for scientific knowledge representation that will enable visual, intuitive, and interactive presentations of complex analytical workflows, while simultaneously simplifying data discovery, accelerating data analysis, building community, and guaranteeing bioinformatic reproducibility.
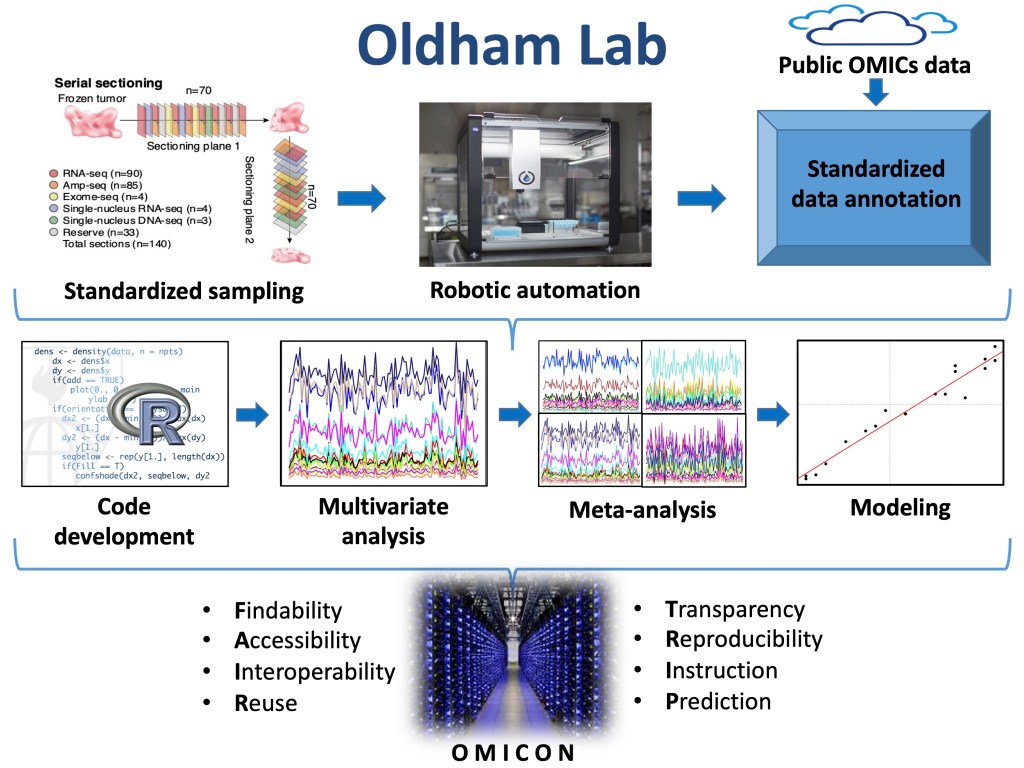
To accelerate efforts to pierce biological complexity and develop new therapies for human diseases, the Oldham Lab seeks to promote integrative, communal, transparent, reproducible, and predictive analyses of biological systems, while standardizing and automating production of gold-standard datasets for this purpose.